The IHMP, also known as Human Microbiome Project 2 (HMP2), is a large-scale initiative designed to explore host–microbiome interplay, including immunity, metabolism, and dynamic molecular activity, to gain a more holistic view of host–microbe interactions over time.
Disease-targeted projects within iHMP comprised pregnancy and preterm birth (PTB); inflammatory bowel diseases (IBD); and stressors that affect individuals with prediabetes.
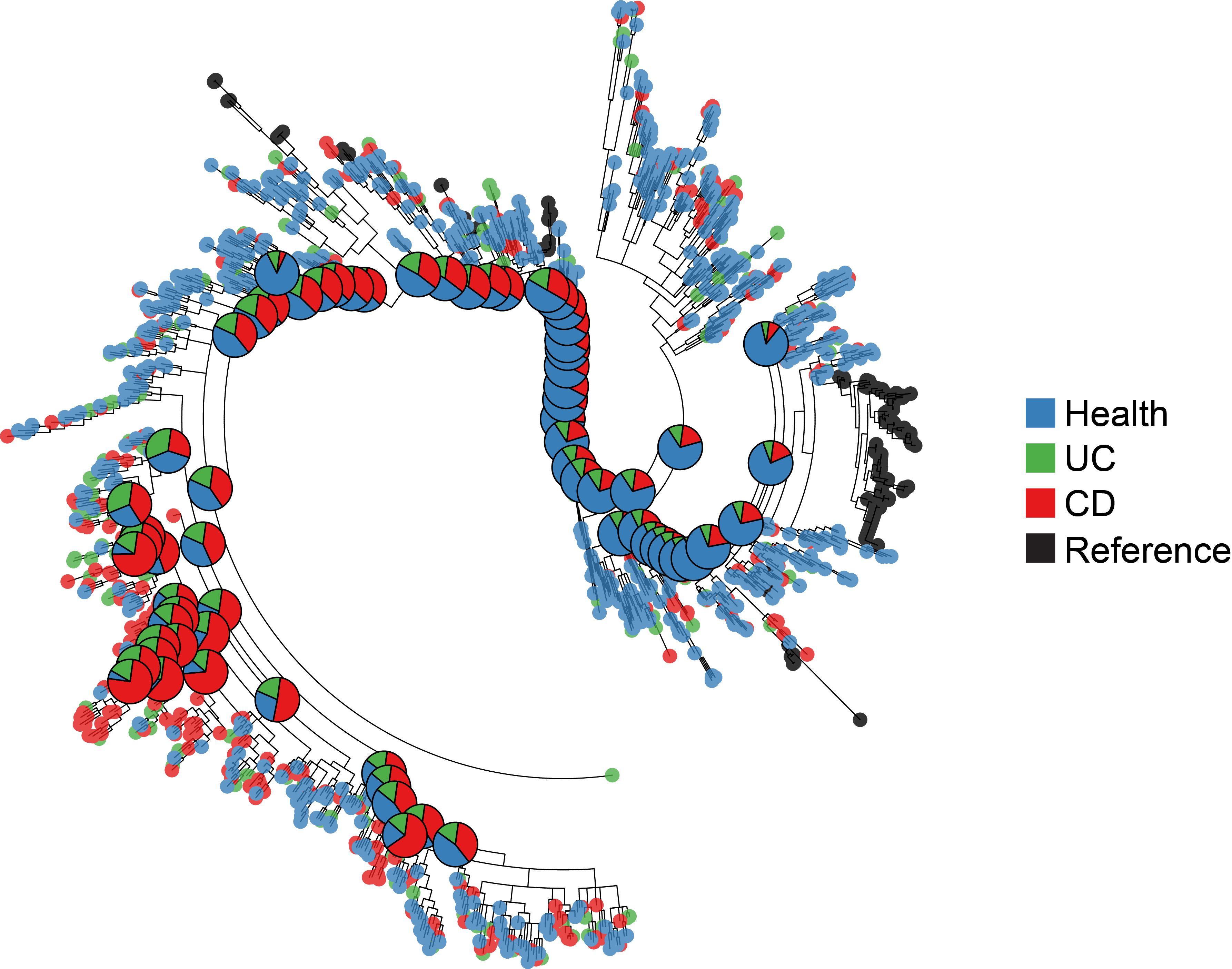
IBD-focused research integrated longitudinal molecular profiles of microbial and host activity by analysing 1,785 stool samples, 651 intestinal biopsies, and 529 quarterly blood samples from 132 individuals and collected over the course of one year each. Key studies led by CSIBD investigators and colleagues can be found below.
Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases
The Inflammatory Bowel Disease Multi'omics Database includes longitudinal data encompassing a multitude of analyses of stool, blood and biopsies of more than 100 individuals, and provides a comprehensive description of host and microbial activities in inflammatory bowel diseases.
Gut microbiome structure and metabolic activity in inflammatory bowel disease
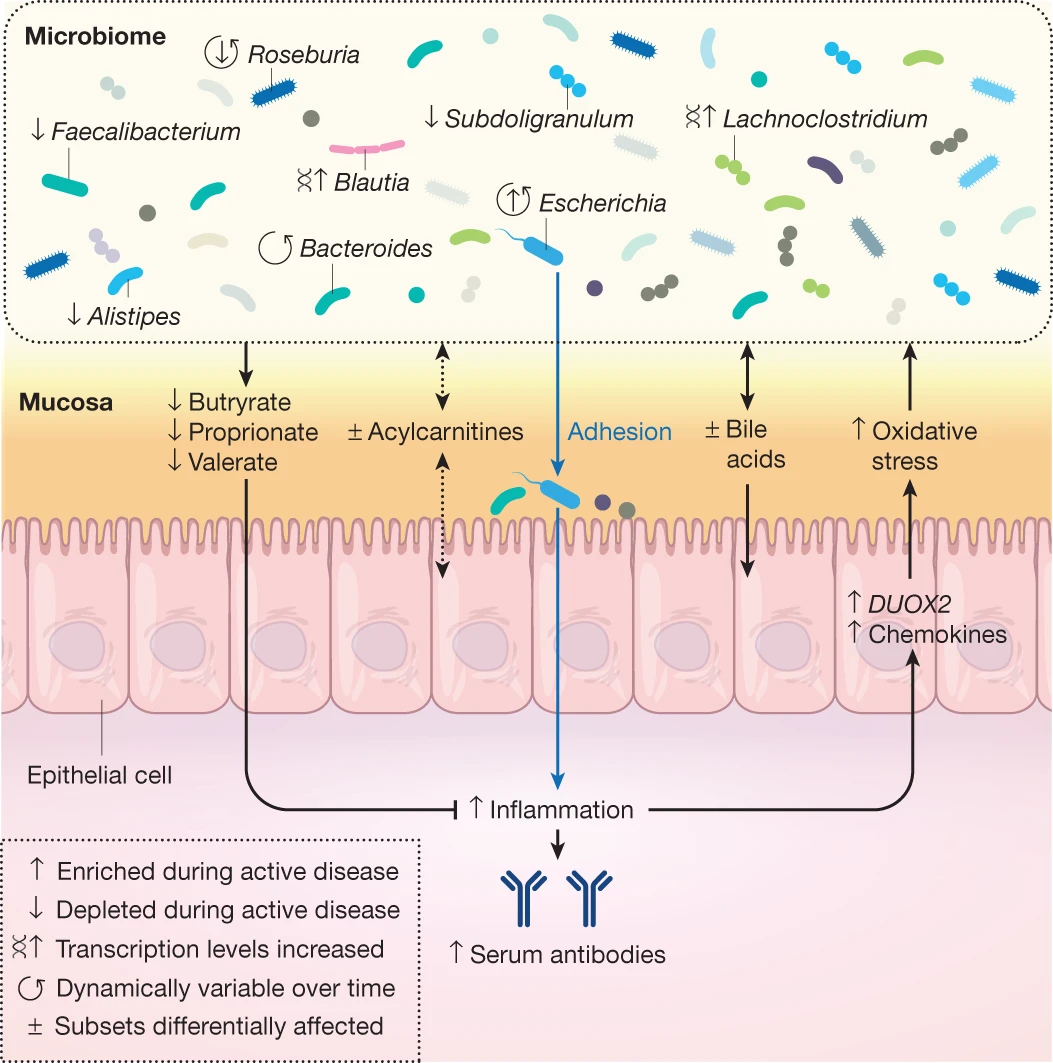
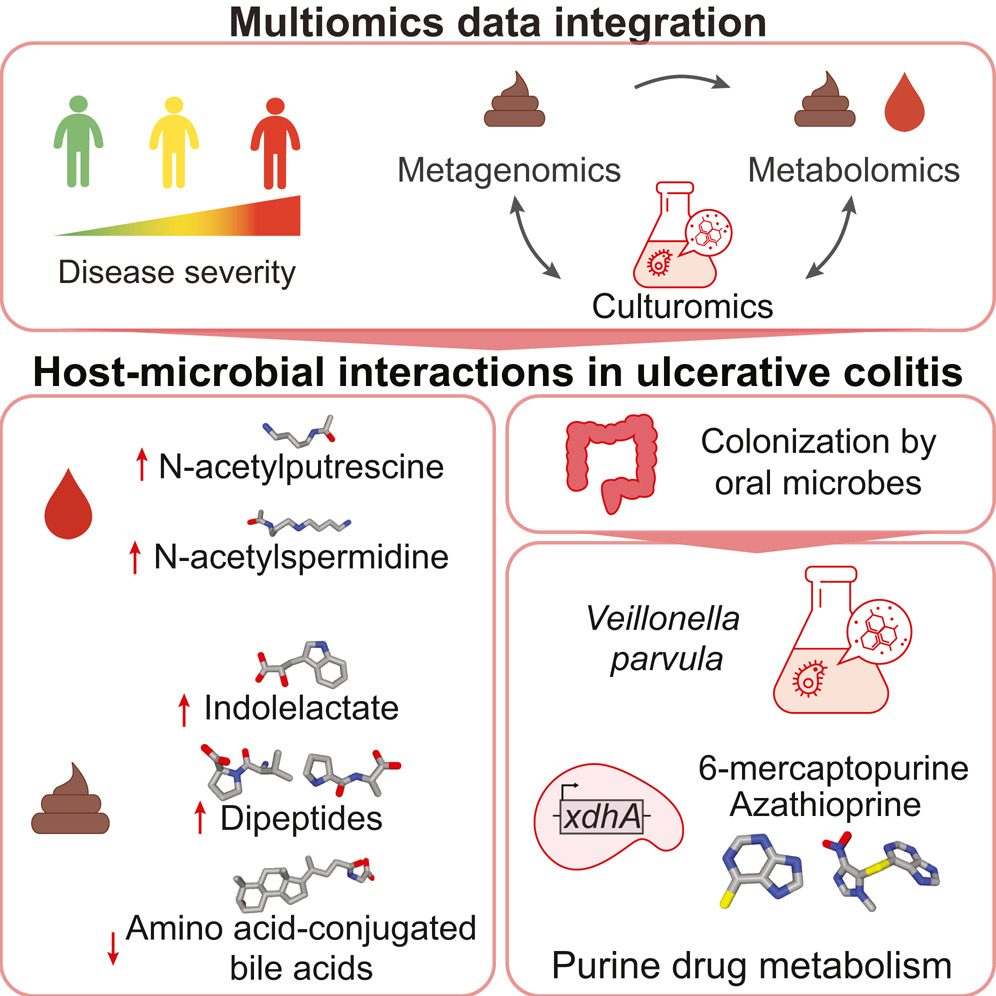
Using metabolomics and shotgun metagenomics on stool samples from individuals with and without inflammatory bowel disease, metabolites, microbial species and genes associated with disease were identified and validated in an independent cohort.
Dynamics of metatranscription in the inflammatory bowel disease gut microbiome
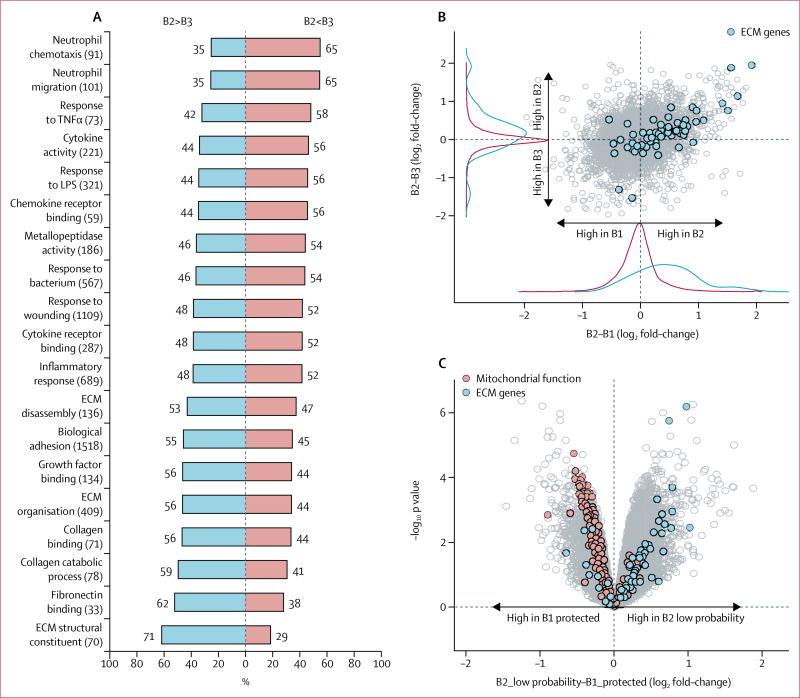
Analysis of paired metagenomes and metatranscriptomes associated with patients with inflammatory bowel disease (IBD) and non-IBD controls over time provides some insights into microbial community variation and potential pathways influencing IBD symptoms.
Strains, functions and dynamics in the expanded Human Microbiome Project
Updates from the Human Microbiome Project analyse the largest known body-wide metagenomic profile of human microbiome personalization.
Structure, function and diversity of the healthy human microbiome
The Human Microbiome Project Consortium reports the first results of their analysis of microbial communities from distinct, clinically relevant body habitats in a human cohort; the insights into the microbial communities of a healthy population lay foundations for future exploration of the epidemiology, ecology and translational applications of the human microbiome.